QM9 Regression#
In this tutorial we provide a 3D example using the qm9-dataset. We use the 3D molecules in the dataset for featurization and train a random forest regressor to predict one of the target properties (here: cv, heat capacity) in the dataset.
To follow along, make sure to install additional dependencies: molflux[rdkit].
Loading the qm9 dataset#
Since the original dataset is quite big, for demonstration reasons, we load a truncated version of the dataset stored on disk
from molflux.datasets import load_dataset_from_store
dataset = load_dataset_from_store("gdb9_trunc.parquet")
print(dataset)
Dataset({
features: ['mol_bytes', 'mol_id', 'A', 'B', 'C', 'mu', 'alpha', 'homo', 'lumo', 'gap', 'r2', 'zpve', 'u0', 'u298', 'h298', 'g298', 'cv', 'u0_atom', 'u298_atom', 'h298_atom', 'g298_atom'],
num_rows: 1000
})
Registering a custom featurization method#
Here we demonstrate on how to add your own representation temporarily. This is a simple wrapper for one of the many 3d-descriptors offered by rdkit descriptors.
from typing import Any, Dict
from molflux.features.catalogue import register_representation
from molflux.features.bases import RepresentationBase
from molflux.features.info import RepresentationInfo
from rdkit import Chem
from rdkit.Chem import Descriptors3D
_DESCRIPTION = """
Rdkit 3D descriptors.
"""
@register_representation(kind="custom", name="rdkit_descriptors_3d")
class RdkitDescriptors3D(RepresentationBase):
def _info(self) -> RepresentationInfo:
return RepresentationInfo(
description=_DESCRIPTION,
)
def _featurise(
self,
samples: Any,
**kwargs: Any,
) -> Dict[str, Any]:
"""compute your features!
Input: a matrix of an array-like representation x N (number of Datapoints in dataset)
"""
samples = [Chem.Mol(mol) for mol in samples]
descriptors = []
for mol in samples:
descriptors.append(getattr(Descriptors3D.rdMolDescriptors, "CalcAUTOCORR3D")(mol))
return {self.tag: descriptors}
Featurising#
Now that we registered the representation, we can load it as if this were part of the catalogue.
from molflux.datasets import featurise_dataset
from molflux.features import load_from_dicts as load_representations_from_dicts
featuriser = load_representations_from_dicts(
[
{"name": "rdkit_descriptors_3d"},
]
)
featurised_dataset = featurise_dataset(
dataset, column="mol_bytes", representations=featuriser
)
In order to compare it to classic molecular descriptors, we create a new column smiles using huggingface datasets mapping functionality.
# Now we can compare to 2D-feature, we need to first generate smiles with a small helper function
def _from_molbytes_to_smiles(example):
mol = Chem.Mol(example["mol_bytes"])
smiles = Chem.MolToSmiles(mol)
# Catch molecules with which rdkit struggles.
mol = Chem.MolFromSmiles(smiles)
if mol is None:
example["smiles"] = None
return example
else:
smiles = Chem.MolToSmiles(mol)
example["smiles"] = smiles
return example
featurised_dataset = featurised_dataset.map(_from_molbytes_to_smiles)
featurised_dataset = featurised_dataset.filter(
lambda example: example["smiles"] is not None
)
featuriser = load_representations_from_dicts(
[
{"name": "morgan"},
{"name": "maccs_rdkit"},
]
)
featurised_dataset = featurise_dataset(
featurised_dataset, column="smiles", representations=featuriser
)
featurised_dataset
Dataset({
features: ['mol_bytes', 'mol_id', 'A', 'B', 'C', 'mu', 'alpha', 'homo', 'lumo', 'gap', 'r2', 'zpve', 'u0', 'u298', 'h298', 'g298', 'cv', 'u0_atom', 'u298_atom', 'h298_atom', 'g298_atom', 'mol_bytes::rdkit_descriptors3_d', 'smiles', 'smiles::morgan', 'smiles::maccs_rdkit'],
num_rows: 991
})
Splitting#
from molflux.datasets import split_dataset
from molflux.splits import load_from_dict as load_split_from_dict
shuffle_strategy = load_split_from_dict(
{
"name": "shuffle_split",
"presets": {
"train_fraction": 0.8,
"validation_fraction": 0.0,
"test_fraction": 0.2,
},
}
)
split_featurised_dataset = next(split_dataset(featurised_dataset, shuffle_strategy))
split_featurised_dataset
DatasetDict({
train: Dataset({
features: ['mol_bytes', 'mol_id', 'A', 'B', 'C', 'mu', 'alpha', 'homo', 'lumo', 'gap', 'r2', 'zpve', 'u0', 'u298', 'h298', 'g298', 'cv', 'u0_atom', 'u298_atom', 'h298_atom', 'g298_atom', 'mol_bytes::rdkit_descriptors3_d', 'smiles', 'smiles::morgan', 'smiles::maccs_rdkit'],
num_rows: 792
})
validation: Dataset({
features: ['mol_bytes', 'mol_id', 'A', 'B', 'C', 'mu', 'alpha', 'homo', 'lumo', 'gap', 'r2', 'zpve', 'u0', 'u298', 'h298', 'g298', 'cv', 'u0_atom', 'u298_atom', 'h298_atom', 'g298_atom', 'mol_bytes::rdkit_descriptors3_d', 'smiles', 'smiles::morgan', 'smiles::maccs_rdkit'],
num_rows: 0
})
test: Dataset({
features: ['mol_bytes', 'mol_id', 'A', 'B', 'C', 'mu', 'alpha', 'homo', 'lumo', 'gap', 'r2', 'zpve', 'u0', 'u298', 'h298', 'g298', 'cv', 'u0_atom', 'u298_atom', 'h298_atom', 'g298_atom', 'mol_bytes::rdkit_descriptors3_d', 'smiles', 'smiles::morgan', 'smiles::maccs_rdkit'],
num_rows: 199
})
})
Training using 3D-descriptors#
import json
from molflux.modelzoo import load_from_dict as load_model_from_dict
from molflux.metrics import load_suite
import matplotlib.pyplot as plt
y_feature = "cv"
model = load_model_from_dict(
{
"name": "random_forest_regressor",
"config": {
"x_features": ["mol_bytes::rdkit_descriptors3_d"],
"y_features": [y_feature],
},
}
)
model.train(split_featurised_dataset["train"])
preds = model.predict(split_featurised_dataset["test"])
regression_suite = load_suite("regression")
scores = regression_suite.compute(
references=split_featurised_dataset["test"][y_feature],
predictions=preds[f"random_forest_regressor::{y_feature}"],
)
print(json.dumps({k: round(v, 2) for k, v in scores.items()}, indent=4))
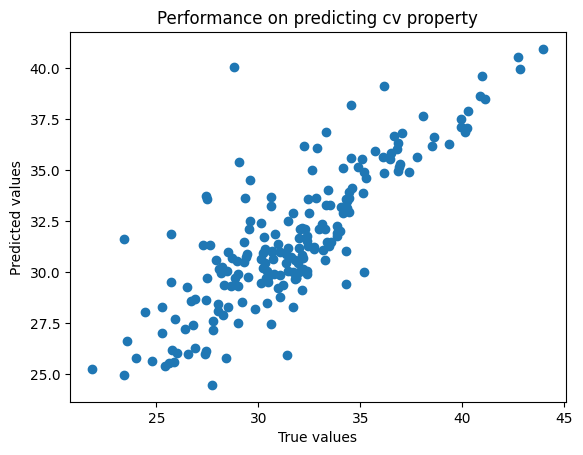
plt.scatter(
split_featurised_dataset["test"][y_feature],
preds[f"random_forest_regressor::{y_feature}"],
)
plt.xlabel("True values")
plt.ylabel("Predicted values")
plt.title(f"Performance on predicting {y_feature} property")
plt.show()
{
"explained_variance": 0.91,
"max_error": 4.21,
"mean_absolute_error": 0.98,
"mean_squared_error": 1.55,
"root_mean_squared_error": 1.24,
"median_absolute_error": 0.81,
"r2": 0.91,
"spearman::correlation": 0.94,
"spearman::p_value": 0.0,
"pearson::correlation": 0.95,
"pearson::p_value": 0.0
}
Training using 2D-descriptors#
model = load_model_from_dict(
{
"name": "random_forest_regressor",
"config": {
"x_features": ["smiles::morgan", "smiles::maccs_rdkit"],
"y_features": [y_feature],
},
}
)
model.train(split_featurised_dataset["train"])
preds = model.predict(split_featurised_dataset["test"])
regression_suite = load_suite("regression")
scores = regression_suite.compute(
references=split_featurised_dataset["test"][y_feature],
predictions=preds[f"random_forest_regressor::{y_feature}"],
)
print(json.dumps({k: round(v, 2) for k, v in scores.items()}, indent=4))
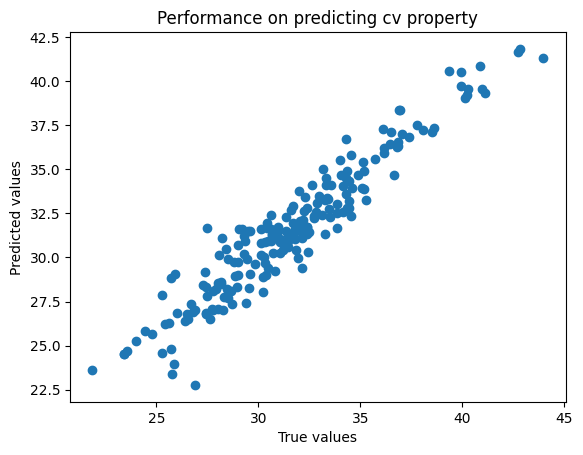
plt.scatter(
split_featurised_dataset["test"][y_feature],
preds[f"random_forest_regressor::{y_feature}"],
)
plt.xlabel("True values")
plt.ylabel("Predicted values")
plt.title(f"Performance on predicting {y_feature} property")
plt.show()
{
"explained_variance": 0.68,
"max_error": 11.26,
"mean_absolute_error": 1.74,
"mean_squared_error": 5.41,
"root_mean_squared_error": 2.33,
"median_absolute_error": 1.45,
"r2": 0.68,
"spearman::correlation": 0.78,
"spearman::p_value": 0.0,
"pearson::correlation": 0.82,
"pearson::p_value": 0.0
}