Energy and forces training#
In this tutorial we provide an example of training physicsml models on both energies and forces from the ani1x
dataset. In this example we choose the nequip_model. We require the rdkitpackage, so make sure to
pip install 'physicsml[rdkit]' to follow along!
Loading the ANI1x dataset#
First, let’s load the ani1x dataset. We will load a truncated version of the dataset (as it’s too large to load
in the docs). For more information on the loading and using dataset, see the molflux documentation.
from molflux.datasets import load_dataset_from_store
dataset = load_dataset_from_store("ani1x_truncated.parquet")
print(dataset)
Dataset({
features: ['mol_bytes', 'chemical_formula', 'ccsd(t)_cbs.energy', 'hf_dz.energy', 'hf_qz.energy', 'hf_tz.energy', 'mp2_dz.corr_energy', 'mp2_qz.corr_energy', 'mp2_tz.corr_energy', 'npno_ccsd(t)_dz.corr_energy', 'npno_ccsd(t)_tz.corr_energy', 'tpno_ccsd(t)_dz.corr_energy', 'wb97x_dz.cm5_charges', 'wb97x_dz.dipole', 'wb97x_dz.energy', 'wb97x_dz.forces', 'wb97x_dz.hirshfeld_charges', 'wb97x_dz.quadrupole', 'wb97x_tz.dipole', 'wb97x_tz.energy', 'wb97x_tz.forces', 'wb97x_tz.mbis_charges', 'wb97x_tz.mbis_dipoles', 'wb97x_tz.mbis_octupoles', 'wb97x_tz.mbis_quadrupoles', 'wb97x_tz.mbis_volumes'],
num_rows: 1000
})
Note
The dataset above is a truncated version to run efficiently in the docs. For running this locally, load the entire dataset by doing
from molflux.datasets import load_dataset
dataset = load_dataset("ani1x", "rdkit")
You can see that there is the mol_bytes column (which is the rdkit serialisation of the 3d molecules) and the
remaining columns of computes properties.
Featurising#
Next, we will featurise the dataset. We extract the atomic numbers, coordinates, and atomic self energies. For more information on the physicsml features, see here.
import logging
logging.disable(logging.CRITICAL)
from molflux.core import featurise_dataset
featurisation_metadata = {
"version": 1,
"config": [
{
"column": "mol_bytes",
"representations": [
{
"name": "physicsml_features",
"config": {
"atomic_number_mapping": {
1: 0,
6: 1,
7: 2,
8: 3,
},
"atomic_energies": {
1: -0.5894385,
6: -38.103158,
7: -54.724035,
8: -75.196441,
},
"backend": "rdkit",
},
"as": "{feature_name}"
}
]
}
]
}
# featurise the mols
featurised_dataset = featurise_dataset(
dataset,
featurisation_metadata=featurisation_metadata,
num_proc=4,
batch_size=100,
)
print(featurised_dataset)
Dataset({
features: ['mol_bytes', 'chemical_formula', 'ccsd(t)_cbs.energy', 'hf_dz.energy', 'hf_qz.energy', 'hf_tz.energy', 'mp2_dz.corr_energy', 'mp2_qz.corr_energy', 'mp2_tz.corr_energy', 'npno_ccsd(t)_dz.corr_energy', 'npno_ccsd(t)_tz.corr_energy', 'tpno_ccsd(t)_dz.corr_energy', 'wb97x_dz.cm5_charges', 'wb97x_dz.dipole', 'wb97x_dz.energy', 'wb97x_dz.forces', 'wb97x_dz.hirshfeld_charges', 'wb97x_dz.quadrupole', 'wb97x_tz.dipole', 'wb97x_tz.energy', 'wb97x_tz.forces', 'wb97x_tz.mbis_charges', 'wb97x_tz.mbis_dipoles', 'wb97x_tz.mbis_octupoles', 'wb97x_tz.mbis_quadrupoles', 'wb97x_tz.mbis_volumes', 'physicsml_atom_idxs', 'physicsml_atom_numbers', 'physicsml_coordinates', 'physicsml_total_atomic_energy'],
num_rows: 1000
})
You can see that we now have the extra columns for
physicsml_atom_idxs: The index of the atoms in the moleculesphysicsml_atom_numbers: The atomic numbers mapped using the mapping dictionaryphysicsml_coordinates: The coordinates of the atomsphysicsml_total_atomic_energy: The total atomic self energy
Splitting#
Next, we need to split the dataset. For this, we use the simple shuffle_split (random split) with 80% training and
20% test. To split the dataset, we use the split_dataset function from molflux.datasets.
from molflux.datasets import split_dataset
from molflux.splits import load_from_dict as load_split_from_dict
shuffle_strategy = load_split_from_dict(
{
"name": "shuffle_split",
"presets": {
"train_fraction": 0.8,
"validation_fraction": 0.0,
"test_fraction": 0.2,
}
}
)
split_featurised_dataset = next(split_dataset(featurised_dataset, shuffle_strategy))
print(split_featurised_dataset)
DatasetDict({
train: Dataset({
features: ['mol_bytes', 'chemical_formula', 'ccsd(t)_cbs.energy', 'hf_dz.energy', 'hf_qz.energy', 'hf_tz.energy', 'mp2_dz.corr_energy', 'mp2_qz.corr_energy', 'mp2_tz.corr_energy', 'npno_ccsd(t)_dz.corr_energy', 'npno_ccsd(t)_tz.corr_energy', 'tpno_ccsd(t)_dz.corr_energy', 'wb97x_dz.cm5_charges', 'wb97x_dz.dipole', 'wb97x_dz.energy', 'wb97x_dz.forces', 'wb97x_dz.hirshfeld_charges', 'wb97x_dz.quadrupole', 'wb97x_tz.dipole', 'wb97x_tz.energy', 'wb97x_tz.forces', 'wb97x_tz.mbis_charges', 'wb97x_tz.mbis_dipoles', 'wb97x_tz.mbis_octupoles', 'wb97x_tz.mbis_quadrupoles', 'wb97x_tz.mbis_volumes', 'physicsml_atom_idxs', 'physicsml_atom_numbers', 'physicsml_coordinates', 'physicsml_total_atomic_energy'],
num_rows: 800
})
validation: Dataset({
features: ['mol_bytes', 'chemical_formula', 'ccsd(t)_cbs.energy', 'hf_dz.energy', 'hf_qz.energy', 'hf_tz.energy', 'mp2_dz.corr_energy', 'mp2_qz.corr_energy', 'mp2_tz.corr_energy', 'npno_ccsd(t)_dz.corr_energy', 'npno_ccsd(t)_tz.corr_energy', 'tpno_ccsd(t)_dz.corr_energy', 'wb97x_dz.cm5_charges', 'wb97x_dz.dipole', 'wb97x_dz.energy', 'wb97x_dz.forces', 'wb97x_dz.hirshfeld_charges', 'wb97x_dz.quadrupole', 'wb97x_tz.dipole', 'wb97x_tz.energy', 'wb97x_tz.forces', 'wb97x_tz.mbis_charges', 'wb97x_tz.mbis_dipoles', 'wb97x_tz.mbis_octupoles', 'wb97x_tz.mbis_quadrupoles', 'wb97x_tz.mbis_volumes', 'physicsml_atom_idxs', 'physicsml_atom_numbers', 'physicsml_coordinates', 'physicsml_total_atomic_energy'],
num_rows: 0
})
test: Dataset({
features: ['mol_bytes', 'chemical_formula', 'ccsd(t)_cbs.energy', 'hf_dz.energy', 'hf_qz.energy', 'hf_tz.energy', 'mp2_dz.corr_energy', 'mp2_qz.corr_energy', 'mp2_tz.corr_energy', 'npno_ccsd(t)_dz.corr_energy', 'npno_ccsd(t)_tz.corr_energy', 'tpno_ccsd(t)_dz.corr_energy', 'wb97x_dz.cm5_charges', 'wb97x_dz.dipole', 'wb97x_dz.energy', 'wb97x_dz.forces', 'wb97x_dz.hirshfeld_charges', 'wb97x_dz.quadrupole', 'wb97x_tz.dipole', 'wb97x_tz.energy', 'wb97x_tz.forces', 'wb97x_tz.mbis_charges', 'wb97x_tz.mbis_dipoles', 'wb97x_tz.mbis_octupoles', 'wb97x_tz.mbis_quadrupoles', 'wb97x_tz.mbis_volumes', 'physicsml_atom_idxs', 'physicsml_atom_numbers', 'physicsml_coordinates', 'physicsml_total_atomic_energy'],
num_rows: 200
})
})
For more information about splitting datasets, see the molflux splitting documentation.
Training the model#
We can now turn to training the model! We choose the nequip_model (or actually a smaller version of it for the example).
To do so, we need to define the model config.
model_config = {
"name": "nequip_model",
"config": {
"x_features": [
'physicsml_atom_idxs',
'physicsml_atom_numbers',
'physicsml_coordinates',
'physicsml_total_atomic_energy',
],
"y_features": [
'wb97x_dz.energy',
'wb97x_dz.forces',
],
"num_node_feats": 4,
"num_features": 5,
"num_layers": 2,
"max_ell": 1,
"compute_forces": True,
"y_graph_scalars_loss_config": {
"name": "MSELoss",
"weight": 1.0,
},
"y_node_vector_loss_config": {
"name": "MSELoss",
"weight": 0.5,
},
"optimizer": {
"name": "AdamW",
"config": {
"lr": 1e-3,
}
},
"scheduler": None,
"datamodule": {
"y_graph_scalars": ['wb97x_dz.energy'],
"y_node_vector": 'wb97x_dz.forces',
"num_elements": 4,
"cut_off": 5.0,
"pre_batch": "in_memory",
"train": {"batch_size": 64},
"validation": {"batch_size": 128},
},
"trainer": {
"max_epochs": 10,
"accelerator": "cpu",
"logger": False,
}
}
}
In the y_features we specify all the columns required for training (energy and forces). In the datamodule, we provide
more details about what each y feature is: energy is a y_graph_scalars and forces are a y_node_vector. We also
specify that the model should compute_forces by computing the gradients of the y_graph_scalars instead of predicting
a y_node_vector directly (which is possible in some models). Finally, we specify two loss configs, one for the
y_graph_scalars_loss_config and one for y_node_vector_loss_config (each of which can be weighted with a weight).
We can now train the model and compute some predictions!
import json
from molflux.modelzoo import load_from_dict as load_model_from_dict
from molflux.metrics import load_suite
import matplotlib.pyplot as plt
model = load_model_from_dict(model_config)
model.train(
train_data=split_featurised_dataset["train"]
)
preds = model.predict(
split_featurised_dataset["test"],
datamodule_config={"predict": {"batch_size": 256}}
)
print(preds.keys())
/home/runner/work/physicsml/physicsml/.cache/nox/docs_build-3-11/lib/python3.11/site-packages/torch/jit/_trace.py:763: TracerWarning: Encountering a list at the output of the tracer might cause the trace to be incorrect, this is only valid if the container structure does not change based on the module's inputs. Consider using a constant container instead (e.g. for `list`, use a `tuple` instead. for `dict`, use a `NamedTuple` instead). If you absolutely need this and know the side effects, pass strict=False to trace() to allow this behavior.
traced = torch._C._create_function_from_trace(
/home/runner/work/physicsml/physicsml/.cache/nox/docs_build-3-11/lib/python3.11/site-packages/torch/jit/_check.py:178: UserWarning: The TorchScript type system doesn't support instance-level annotations on empty non-base types in `__init__`. Instead, either 1) use a type annotation in the class body, or 2) wrap the type in `torch.jit.Attribute`.
warnings.warn(
/home/runner/work/physicsml/physicsml/.cache/nox/docs_build-3-11/lib/python3.11/site-packages/torch/jit/_check.py:178: UserWarning: The TorchScript type system doesn't support instance-level annotations on empty non-base types in `__init__`. Instead, either 1) use a type annotation in the class body, or 2) wrap the type in `torch.jit.Attribute`.
warnings.warn(
/home/runner/work/physicsml/physicsml/.cache/nox/docs_build-3-11/lib/python3.11/site-packages/torch/jit/_check.py:178: UserWarning: The TorchScript type system doesn't support instance-level annotations on empty non-base types in `__init__`. Instead, either 1) use a type annotation in the class body, or 2) wrap the type in `torch.jit.Attribute`.
warnings.warn(
/home/runner/work/physicsml/physicsml/.cache/nox/docs_build-3-11/lib/python3.11/site-packages/torch/jit/_check.py:178: UserWarning: The TorchScript type system doesn't support instance-level annotations on empty non-base types in `__init__`. Instead, either 1) use a type annotation in the class body, or 2) wrap the type in `torch.jit.Attribute`.
warnings.warn(
/home/runner/work/physicsml/physicsml/.cache/nox/docs_build-3-11/lib/python3.11/site-packages/torch/jit/_check.py:178: UserWarning: The TorchScript type system doesn't support instance-level annotations on empty non-base types in `__init__`. Instead, either 1) use a type annotation in the class body, or 2) wrap the type in `torch.jit.Attribute`.
warnings.warn(
/home/runner/work/physicsml/physicsml/.cache/nox/docs_build-3-11/lib/python3.11/site-packages/torch/jit/_check.py:178: UserWarning: The TorchScript type system doesn't support instance-level annotations on empty non-base types in `__init__`. Instead, either 1) use a type annotation in the class body, or 2) wrap the type in `torch.jit.Attribute`.
warnings.warn(
/home/runner/work/physicsml/physicsml/.cache/nox/docs_build-3-11/lib/python3.11/site-packages/torch/jit/_check.py:178: UserWarning: The TorchScript type system doesn't support instance-level annotations on empty non-base types in `__init__`. Instead, either 1) use a type annotation in the class body, or 2) wrap the type in `torch.jit.Attribute`.
warnings.warn(
/home/runner/work/physicsml/physicsml/.cache/nox/docs_build-3-11/lib/python3.11/site-packages/torch/jit/_check.py:178: UserWarning: The TorchScript type system doesn't support instance-level annotations on empty non-base types in `__init__`. Instead, either 1) use a type annotation in the class body, or 2) wrap the type in `torch.jit.Attribute`.
warnings.warn(
/home/runner/work/physicsml/physicsml/.cache/nox/docs_build-3-11/lib/python3.11/site-packages/torch/jit/_check.py:178: UserWarning: The TorchScript type system doesn't support instance-level annotations on empty non-base types in `__init__`. Instead, either 1) use a type annotation in the class body, or 2) wrap the type in `torch.jit.Attribute`.
warnings.warn(
/home/runner/work/physicsml/physicsml/.cache/nox/docs_build-3-11/lib/python3.11/site-packages/torch/jit/_check.py:178: UserWarning: The TorchScript type system doesn't support instance-level annotations on empty non-base types in `__init__`. Instead, either 1) use a type annotation in the class body, or 2) wrap the type in `torch.jit.Attribute`.
warnings.warn(
/home/runner/work/physicsml/physicsml/.cache/nox/docs_build-3-11/lib/python3.11/site-packages/torch/jit/_check.py:178: UserWarning: The TorchScript type system doesn't support instance-level annotations on empty non-base types in `__init__`. Instead, either 1) use a type annotation in the class body, or 2) wrap the type in `torch.jit.Attribute`.
warnings.warn(
/home/runner/work/physicsml/physicsml/.cache/nox/docs_build-3-11/lib/python3.11/site-packages/torch/jit/_check.py:178: UserWarning: The TorchScript type system doesn't support instance-level annotations on empty non-base types in `__init__`. Instead, either 1) use a type annotation in the class body, or 2) wrap the type in `torch.jit.Attribute`.
warnings.warn(
/home/runner/work/physicsml/physicsml/.cache/nox/docs_build-3-11/lib/python3.11/site-packages/torch/jit/_check.py:178: UserWarning: The TorchScript type system doesn't support instance-level annotations on empty non-base types in `__init__`. Instead, either 1) use a type annotation in the class body, or 2) wrap the type in `torch.jit.Attribute`.
warnings.warn(
/home/runner/work/physicsml/physicsml/.cache/nox/docs_build-3-11/lib/python3.11/site-packages/torch/jit/_check.py:178: UserWarning: The TorchScript type system doesn't support instance-level annotations on empty non-base types in `__init__`. Instead, either 1) use a type annotation in the class body, or 2) wrap the type in `torch.jit.Attribute`.
warnings.warn(
/home/runner/work/physicsml/physicsml/.cache/nox/docs_build-3-11/lib/python3.11/site-packages/torch/jit/_check.py:178: UserWarning: The TorchScript type system doesn't support instance-level annotations on empty non-base types in `__init__`. Instead, either 1) use a type annotation in the class body, or 2) wrap the type in `torch.jit.Attribute`.
warnings.warn(
/home/runner/work/physicsml/physicsml/.cache/nox/docs_build-3-11/lib/python3.11/site-packages/lightning/pytorch/utilities/data.py:105: Total length of `dict` across ranks is zero. Please make sure this was your intention.
/home/runner/work/physicsml/physicsml/.cache/nox/docs_build-3-11/lib/python3.11/site-packages/physicsml/lightning/pre_batching_in_memory.py:53: You are using `torch.load` with `weights_only=False` (the current default value), which uses the default pickle module implicitly. It is possible to construct malicious pickle data which will execute arbitrary code during unpickling (See https://github.com/pytorch/pytorch/blob/main/SECURITY.md#untrusted-models for more details). In a future release, the default value for `weights_only` will be flipped to `True`. This limits the functions that could be executed during unpickling. Arbitrary objects will no longer be allowed to be loaded via this mode unless they are explicitly allowlisted by the user via `torch.serialization.add_safe_globals`. We recommend you start setting `weights_only=True` for any use case where you don't have full control of the loaded file. Please open an issue on GitHub for any issues related to this experimental feature.
/home/runner/work/physicsml/physicsml/.cache/nox/docs_build-3-11/lib/python3.11/site-packages/lightning/pytorch/trainer/connectors/data_connector.py:424: The 'train_dataloader' does not have many workers which may be a bottleneck. Consider increasing the value of the `num_workers` argument` to `num_workers=3` in the `DataLoader` to improve performance.
/home/runner/work/physicsml/physicsml/.cache/nox/docs_build-3-11/lib/python3.11/site-packages/torch/storage.py:414: FutureWarning: You are using `torch.load` with `weights_only=False` (the current default value), which uses the default pickle module implicitly. It is possible to construct malicious pickle data which will execute arbitrary code during unpickling (See https://github.com/pytorch/pytorch/blob/main/SECURITY.md#untrusted-models for more details). In a future release, the default value for `weights_only` will be flipped to `True`. This limits the functions that could be executed during unpickling. Arbitrary objects will no longer be allowed to be loaded via this mode unless they are explicitly allowlisted by the user via `torch.serialization.add_safe_globals`. We recommend you start setting `weights_only=True` for any use case where you don't have full control of the loaded file. Please open an issue on GitHub for any issues related to this experimental feature.
return torch.load(io.BytesIO(b))
/home/runner/work/physicsml/physicsml/.cache/nox/docs_build-3-11/lib/python3.11/site-packages/lightning/pytorch/core/module.py:516: You called `self.log('train/total/y_node_vector', ..., logger=True)` but have no logger configured. You can enable one by doing `Trainer(logger=ALogger(...))`
/home/runner/work/physicsml/physicsml/.cache/nox/docs_build-3-11/lib/python3.11/site-packages/lightning/pytorch/core/module.py:516: You called `self.log('train/total/y_graph_scalars', ..., logger=True)` but have no logger configured. You can enable one by doing `Trainer(logger=ALogger(...))`
/home/runner/work/physicsml/physicsml/.cache/nox/docs_build-3-11/lib/python3.11/site-packages/lightning/pytorch/core/module.py:516: You called `self.log('train/total/loss', ..., logger=True)` but have no logger configured. You can enable one by doing `Trainer(logger=ALogger(...))`
/home/runner/work/physicsml/physicsml/.cache/nox/docs_build-3-11/lib/python3.11/site-packages/lightning/pytorch/trainer/connectors/data_connector.py:424: The 'predict_dataloader' does not have many workers which may be a bottleneck. Consider increasing the value of the `num_workers` argument` to `num_workers=3` in the `DataLoader` to improve performance.
dict_keys(['nequip_model::wb97x_dz.energy', 'nequip_model::wb97x_dz.forces'])
As you can see the predictions include an energy prediction and a forces prediction. Finally, we compute some metrics
regression_suite = load_suite("regression")
scores = regression_suite.compute(
references=split_featurised_dataset["test"]["wb97x_dz.energy"],
predictions=preds["nequip_model::wb97x_dz.energy"],
)
print(json.dumps(scores, indent=4))
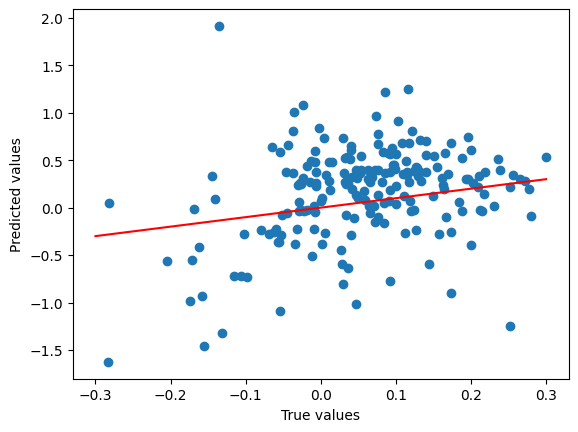
true_shifted = [x - e for x, e in zip(split_featurised_dataset["test"]["wb97x_dz.energy"], split_featurised_dataset["test"]["physicsml_total_atomic_energy"])]
pred_shifted = [x - e for x, e in zip(preds["nequip_model::wb97x_dz.energy"], split_featurised_dataset["test"]["physicsml_total_atomic_energy"])]
plt.scatter(
true_shifted,
pred_shifted,
)
plt.plot([-0.3, 0.3], [-0.3, 0.3], c='r')
plt.xlabel("True values")
plt.ylabel("Predicted values")
plt.show()
{
"explained_variance": 0.9999914063806031,
"max_error": 2.05029296875,
"mean_absolute_error": 0.3690852355957031,
"mean_squared_error": 0.23329730607336388,
"root_mean_squared_error": 0.4830085983430977,
"median_absolute_error": 0.2854766845703125,
"r2": 0.9999910220496709,
"spearman::correlation": 0.9999324983124579,
"spearman::p_value": 0.0,
"pearson::correlation": 0.9999957844008471,
"pearson::p_value": 0.0
}